Moorhen Tutorial 1: Fix up the Cyclin-Dependent Kinase
Welcome to Moorhen (“Coot on the Web”).
Getting Started
There are two ways you can access Moorhen. One is using it within CCP4 Cloud the other one is using it as a stand-alone app at moorhen.org. While both versions are broadly similar, this tutorial assumes that you make use of CCP4 Cloud version as it offers capabilities beyond the scope of Moorhen.
Open a web browser window and point it at cloud.ccp4.ac.uk, then login using your username and password. Once you are in the project folder, click on the “Tutorials” button on the top right. This will open a dialog with a file browser. To reach the hop-on project for this tutorial, follow this path:
- 4. Refinement → T4_006. Fix up the Cyclin-Dependent Kinase with Moorhen
Now open the imported project. This imported project consists of a model for a Kinase Inhibitor together with its experimental data.
Click on “Add Job” (green arrow) and create a new “Model Building with WebCoot/Moorhen” task right at the bottom of the task tree. Click on “Run”.
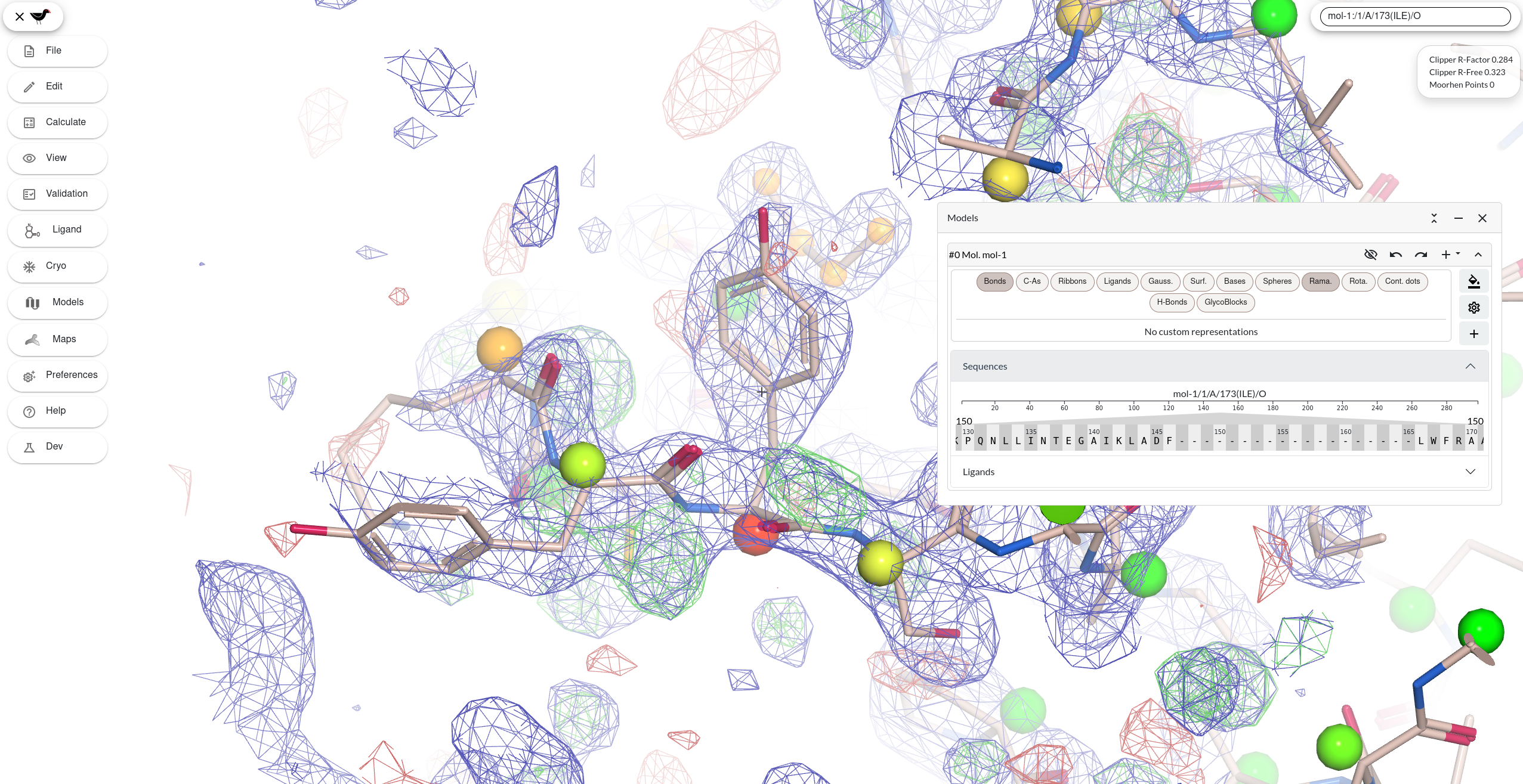
A new window with Moorhen opens. After the initial load, Moorhen displays a protein model, a blue 2Fo-Fc-style map and an Fo-Fc-style map in green (positive) and red (negative).
-
If the new window is too small, you can resize it by pulling its borders or by clicking the “maximize” button ont he top right.
-
Use Left-Mouse click and drag to rotate the view. (just click and drag, when using at trackpad)
-
Use scroll-wheel scroll to zoom in and out (use 2-finger drag on a trackpad)
-
Use middle-mouse click to centre on an atom (use Option-click on a trackpad on a Mac, or Alt-click on a trackpad on a Windows)
-
To pan the view, use middle-mouse click and drag (use Shift Option click and drag on a trackpad on a Mac, or Shift Alt click and drag on a trackpad on a Windows)
You can change the speed that moving the mouse spins the view using the “Preferences” menu:
- First, click on the Moorhen icon on the top left to open the dropdown burger menu.
- Click on Preferences → “Mouse Sensitivity” → 0.4 (for example).
- Click outside the Preferences dialog to make the dialog disappear.
- If you wish to close the burger menu, click again on the Moorhen icon which now displays a cross.
Similarly, you can change the thickness of the map lines if you wish.
- Use “[” and “]” on the keyboard to adjust the radius of the density.
- Ctrl middle-mouse scroll to change the contour level (one step at time).
- Alternatively you can click open the dropdown menu again and click on the “Maps” button. This will show a draggable window where you can use sliders to adjust contour level and radius for each map.
Let’s Go!
Our job is to fix and amend the protein model in a way that is consistent with the data.
Maps and Models
- Open the dropdown burger menu and click on Models.
Moorhen displays a draggable window with a list of models
Initially the background of the window is semi-transparent,
- Put your cursor over the window to make it opaque. This behaviour can be switched off in Preferences.
You will see a dropdown menu labelled “Sequences”. Click on it to open the accordion and reveal the sequence viewer - let’s use that to move around the structure. Click on a few letters (that represent the residues in the protein). Notice that the map density mesh is redrawn around the new centre.
See the grey rectangle over the sequence numbers?
- Move your cursor inside the box and use left-mouse click and drag to move around the sequence.
You can resize the rectangle to display more residue letters (by clicking and dragging on the edge of the box), but if you make it too wide it will not display any.
- Click on the Ligands dropdown tab to open the ligands accordion
Notice that there are “No ligands.” We will add one later.
For higher-end computers we can use a smoother representation of the bonds and atoms.
- [Optional] Click on the gear icon, and change the Bond Smoothness to Smooth.
You can also control the way maps are displayed using the Maps window. Click on the Maps button on the dropdown burger menu.
Moorhen displays a draggable window with a list of maps
You will now see the cards for the maps (the 2Fo-Fc-style map has a blue icon and the difference map has an icon with red and green). You can use the sliders there to adjust the contour level. Clicking (click and release) on the slider is sometimes a better way to adjust the sliders than trying to “slide” the slider using click and drag. Using the +/- buttons on each side of the slider is also a nice way to do fine adjustments.
- You can also use Ctrl scroll-wheel scroll to change the contour level of the map marked as Active.
You can now dismiss the draggable Maps and Models windows using the close button. Alternatively, you may prefer to minimise them so that you don’t have to open and close them multiple times.
Validation Tools
Let’s now look at the Ramachandran plot:
- Click on the Validation button under the dropdown burger menu.
Moorhen displays an empty draggable window with a selector
- Choose Ramachandran Plot in the Tool option menu.
Moorhen shows the Ramachandran Plot for the “A” chain of this protein
You will see that there are several interesting red spots.
- Let’s click on the red spot at the middle top
Moorhen will put residue A180 at the centre of the screen
Take a look at the region… (You can minimise the validation window for a better look).
Hmmm… the carbonyl oxygen atoms are a bit close… Are there any other Ramachandran outliers in the area?
-
To measure the distance between atoms, hold M and click on an atom, then hold M again and click on another atom. Press C to clear all atom labels and measurements.
-
On the Models window, click on the Rama button for this protein model.
Moorhen displays Ramachandran balls that represent the probability of those phi, psi angles for that residue
Residue A180 has a red ball and just upstream at A178 the LYS has a orange ball. Maybe both of these can be improved.
Notice that there is a green blob close to the N of A180. Maybe it would be better that were fitted by the carbonyl of the peptide. Let’s try that:
Flipping a Peptide
- Right click on any atom of the residue A179. Moorhen uses transparent golden balls to let you know which will be the picked atom or residue when you hover over it. There’s a text box on the top right that will also appear letting you know the exact atom selection.
Moorhen displays a context menu consisting of a toolkit with icons for modelling (with which you may already be familiar if you have used Coot)
As you move the mouse over the icons in the toolkit, you will see a tooltip for that icon on the bottom of this context menu.
- To flip the peptide you want to use the “Flip Peptide” button. Click it.
Moorhen flips the Peptide and the Ramachandran ball for that residues turns green. The toolkit dissapears when everything is done
Yay. Progress. Let’s see if we can do the same for residue A178.
- Right click on residue A177, then click on the “Flip Peptide” button again.
Moorhen flips the peptide and the Ramachandran ball for 177 turrns green
More progress. Good stuff.
- Look at the Ramachandran Plot in the draw. Notice that the red spots for the problematic residues have disappeared.
Navigation tip: Use the middle mouse button click over an atom to put the clicked atom at the centre of the screen. If you are using a trackpad, you can also hold Alt while clicking on an atom to achieve the same result.
Real Space Refinement
- Right click residue A178, and then on the “Refine Residues” button (the icon is a bullseye). A selection menu will appear. Choose “Sphere” in this option menu, which means that a set of residues within a fixed radius of the central chosen atom will be refined.
Moorhen refines the sphere of residues
Now the local backbone fits quite nicely into the blue map.
Connect the Maps: Updating Maps
Wouldn’t it be nice though, if the difference map updated, so that if you add atoms to green blobs or removed them from red blobs then those blobs would disappear?
Let’s try that:
- Oepn the dropdown burger menu and then click on File → Connect molecule and map for updating… → OK. (No need to change the values in the options menus because they are already setup to be correct.)
Moorhen will display a “toast” top right informing you of the current R-factor and the number of Moorhen points that you have collected (so far none, because we have just started)
Note: Using updating/connected maps will slow down the model-building process somewhat but we now have the advantage of collection Moorhen points and watching the R-factor go down as we make changes. Moorhen points indicate progress in flattening the difference map.
Difference Map Peaks
Note: Here I find it useful to adjust the map contour level to 0.64 (and 0.47 for the difference map).
If you have closed the validation window, open it again.
- Choose Difference Map Peaks as the validation tool.
Moorhen displays the difference map peaks in a waterfall plot
- Use the slider to change the RMSD to about 5.0.
On the left of the waterfall plot are the most positive peaks (and if there were any the most negative peaks would be displayed on the far right). Click on the biggest/leftmost green peak.
What are we looking at? An orange Ramachandran ball?
Flipping… flipping
It’s a flipped peptide.
- So let’s flip it to the correct orientation using right click on the residue and then choosing Flip peptide.
As we do so, Moorhen makes several updates. It moves the model, it updates the maps in the light of the new model, it updates the Ramachandran balls so that they are both green now. And, in the toast, it updates the R-factor (a tiny amount) and gives us some Moorhen points.
Flipping a peptide to the correct orientation generally gives you 15-20 Moorhen points.
You will notice that that the Difference Map Peaks graph has been updated too - the leftmost peak has gone. Have a look at the next 12 or so peaks. What do you notice?
Adding Waters
You will notice that they are mostly peaks of waters. We could add waters one by one, but a more automated method is to do many at the same time.
- Under the dropdown burger menu, click on Ligand → Add waters… → OK
This will add around 100 waters. And as above, the maps and the R-factors will update and we will get many Moorhen points. The map should improve a bit so that the ligand is more easy to make out.
Contact Dots and Clashes
- Use the sequence viewer on the Models window to navigate to residue A194.
What this? It’s a flipped peptide - let’s flip it back to where it should be. But having done that, what do you notice? Let’s use Moorhen’s clash analysis:
- In the model molecule card for the tutorial structure you will see a box labelled Cont. - click it.
Moorhen display contact dots and clash interactions
Wooh! Pink sticks. Bad news! So let’s also flip the pepide on the next rung of the helix - residue A197.
The contact dots between A194 and A197 disappear
- OK, you can turn off the contact dots for now by unclicking the Cont. box in the molecule card.
Change the Residue Type
Now navigate to residues A193. What do the maps tell you is going on here? What is the residue type? What does the model say? What does the map say?
OK, so first let’s fill the side-chain with the atoms of the type from the main-chain atoms: “TYR” - do right click on residue A193 and then click on Auto-fit Rotamer.
What do we see? What does that suggest?
It suggests that the sequence of the model doesn’t match the sequence of the protein from which the data were collected. OK, so let’s mutate it.
- Again do right click on residue A193. Now click on the Mutate button, then from the residues type chooser currently “ALA (A)” choose “PHE (F)”.
Moorhen updates the map so that the red blob goes away
- Navigate to residue A168.
What do we see? What should it be instead?
Mutate
OK, so let’s mutate it:
- Right click on residue A168 and use Mutate, change the type to “TYR (Y)”.
(More Moorhen Points - yay).
- Let’s go back to the Difference Map Peaks in the Validation tools
Now you can see negative peaks. Let’s have a look at those.
Can you find a negative difference map peak that is close to resiude A187? Have a look at the model? What needs to be changed?
Rotamers
In the Models window, click the box for “Rota”
Moorhen displays rotamer dodecahedrons coloured by rotamer probability
-
So let’s change the rotamer. First right click on residue A187 and then use the Auto-fit Rotamer button.
-
If the side-chain doesn’t lie flat in its density, you can use the Refine Residues in the context menu that opens when you right click on residue A187 to give it a bit of Real Space Refinement that will sort of the fitting.
On improvement of the rotamer probability, Moorhen will change the colour of the dodecahedron to be more green
Can you find a negative difference map peak close to residue A141? Again examine. What do we need to do? Let’s fix the rotamer then using “Auto-fit Rotamer” as before.
Fit the Ligand
OK, now it’s time to fit the ligand!
- Use the Difference Map Peaks to navigate to the ligand.
Several of the top 5 peaks should now correspond to the ligand.
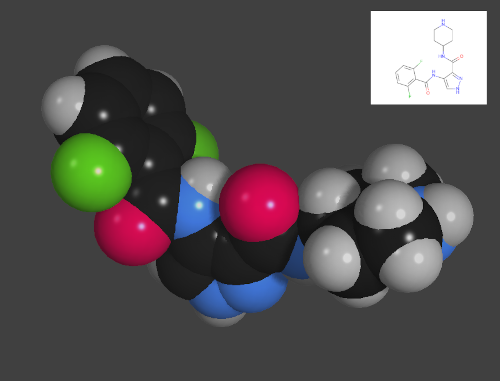
- Ligand → Get Monomer → LZA → OK
Moorhen imports the LZA ligand
OK, fine. Now let’s undisplay it:
- In Models window scroll to the bottom. There should now be a card for the newly imported ligand (“#3 Mol LZA”). Click on the eye icon to undisplay the ligand
Moorhen changes the icon to an uncrossed eye and the ligand disappears
-
On the dropdown burger menu, click on Ligand → Fit ligand here…
-
Change the option menu labelled “Ligand molecule” so that it reads “3: LZA” then click OK.
Moorhen fits the ligand in the local blob
It should be reasonably close but not exact because the algorithm didn’t use conformational variation.
Merge the Ligand
So let’s add the ligand to the protein model:
- On the dropdown burger menu, click on Edit → Merge molecules…
- Change the entry in the option menu labelled “From molecule” so that it reads “4: lig_4”
- Click OK.
Moorhen updates the maps so that the difference map blobs change
Now let’s refine the ligand and the surrounding residues:
- Right click on the ligand and then click on Refine Residues. Change the option menu to “SPHERE” then click on an atom in the ligand
Moorhen updates the maps so that there are no difference maps peaks left on the ligand
Add a Water
- Navigate to a water peak using middle-mouse click and drag (Shift-Option click and drag on a trackpad on a Mac, or Shift-Alt click and drag on a trackpad on a Windows) to pan the view. the view to the water blob.
- You can also use the arrow-keys to pan the view
Let’s add a water here
- On the dropdown burger menu, click on Edit → Add simple
- Change the option menu to read “HOH”
- Click OK
Moorhen adds a water at the centre of the screen
There are several water peaks in the map similar to this.
At some stage, when you add a water, you will see a the contours of negative density over part or all the water peak. What does that mean?
## More Validation Tools
- Open the Validation Tools window
- Click on Validation Plot
Moorhen displays interactive validation graphs.
Over to You!
- Click on the validation bars to navigate to interesting parts of the structure and make some fixes.
You should be able to collect about 1800 Moorhen points. Maybe more!
Make a Pretty Picture
- Using the cards in the Maps window, undisplay the maps using the eye icon
- In the Models window, click on “Bonds” to undisplay the “Bonds” representation of the model
- Likewise undisplay the Rama ball and Rota dodecs if you have the displayed
- Click on Lig. to display the ligand in the model
- Click on Ribb. to display the model in Ribbon mode
To navigate to the ligand:
- On the dropdown burger menu, click on Ligand → Centre on ligand…
- Click “mol-1”
- Click the “/1/A/1(LZA)” label
- On the dropdown burger menu, click on View → Set background colour - change it if you wish
- View → Clipping and fogging…
- Adjust the sliders to make the ligand more clearly visible
- OK
Use keyboard “S” to activate the “in application” screen capture.
Export Your Molecule
There’s no need to save the model into cloud, everything will be saved automatically once we close Moorhen. When you are done, either click on the close button on the top right, or click on Exit under the dropdown burger menu.
A window will open with a message letting you know that the model was saved in cloud and Moorhen will close.
Let’s see if we managed to improve the Rfree using Refmac. In CCP4 Cloud, add a “Refinement using Refmac” task to the tree right under the Moorhen task. Make sure to set the “Number of refinement cycles” to zero and wait for the results. Now do the same, but create the Refmac task at the same level as the Moorhen task, i.e. using the original unmodified model as input. According to Refmac, did the Rfactor and Rfree improve tinkering with the model using Moorhen?
If you are doing this tutorial in moorhen.org then you can export models by clicking on the download button on the top right of the molecule card found in the Models window.
Notes:
The tutorial is based on 2vtq “Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design” Wyatt, P.G. et al. (2008), J. Med. Chem. 51, 4986.